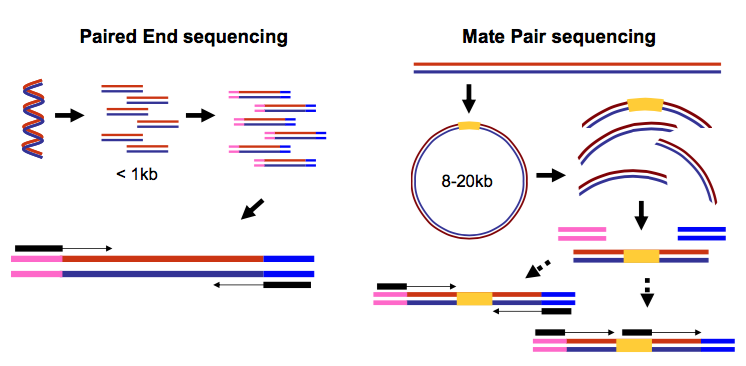
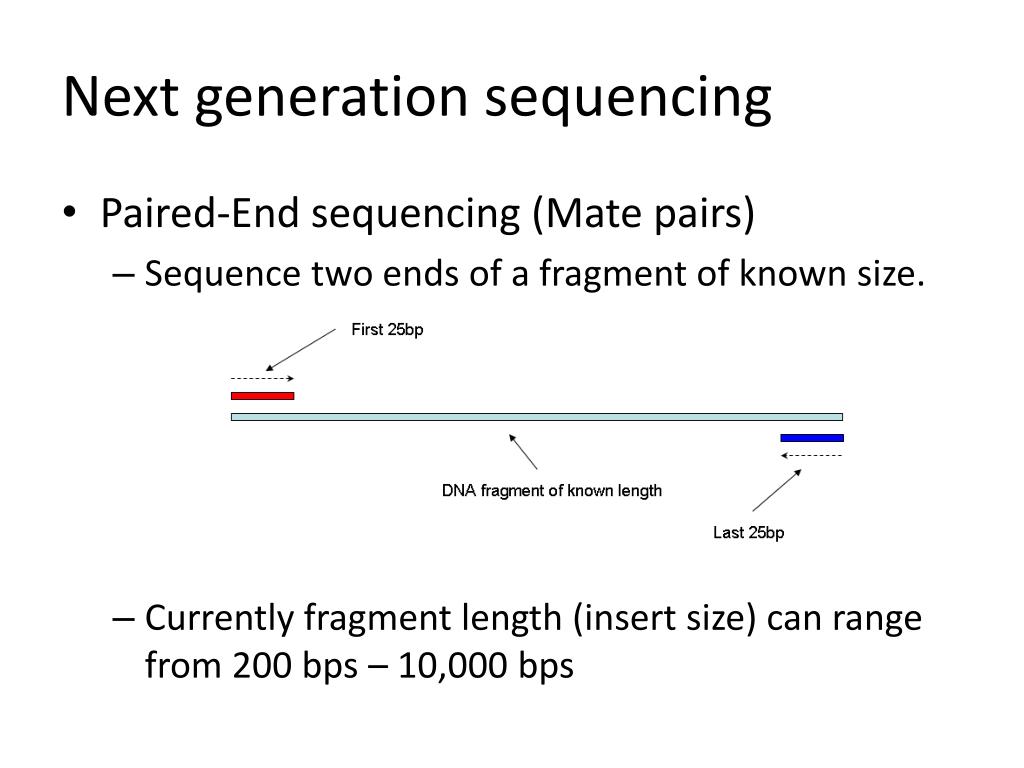
paired end sequencing insert size
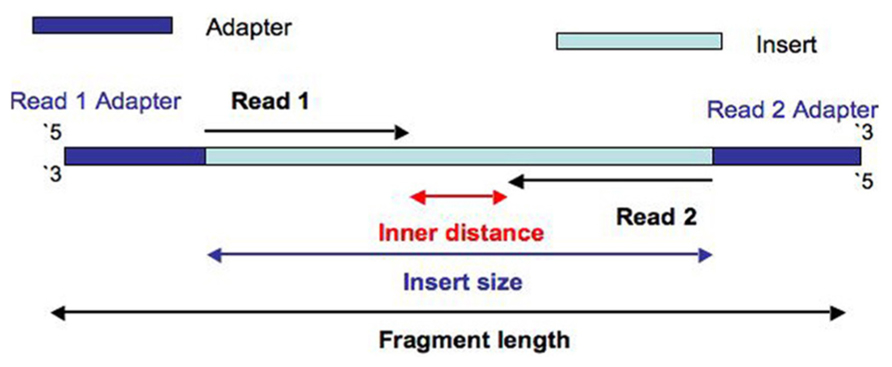
Read files from paired-end sequencing are required to be paired prior to assembly and this can be done using the Set merge paired reads operation. The insert is normally the stretch of sequence between the paired-end adapters so in your case the insert size would be 250 bp 2x75 bp reads 100 bp unsequenced middle piece.
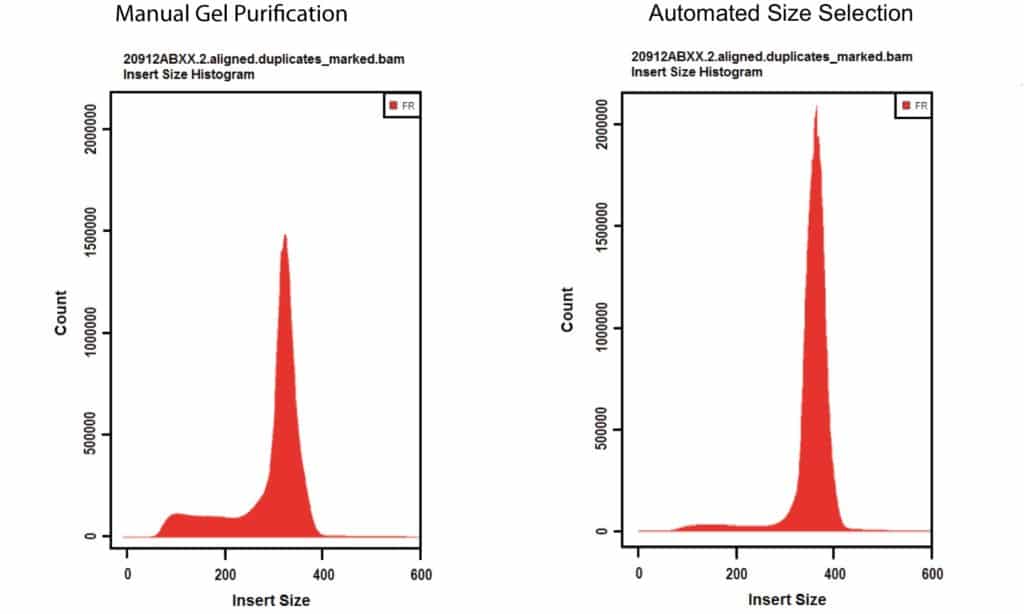
Why Dna Size Selection Matters In Ngs Pipelines
Note that the paired distance you set does not need to be exact for all reads.

. 127649175-1276418 148 This one makes sense to me as we get the total. Including flanking intronic regions of. What is insert size paired end.
To set paired reads. - hello there i read this article httpbioinformaticsUconn. What is the optimal insert size.
Example 1 - inferred insert size is rightmost coordinate read length - leftmost coordinate. The insert size on classic paired-end is smaller about 500bp while the insert size of mate-pair is much longer several Kb which allows to join the contiguous between them. Despite the benefits of gaining SS-PE data with paired ends of varying distance the standard Illumina protocol allows only non-strand-specific paired-end sequencing with a single insert.
It can be useful to produce fragments with an insert. For a brief primer on paired-end sequencing and mate-pair reads see the GATK Dictionary. Collect metrics about the insert size distribution of a paired-end library.
Despite the benefits of gaining SS-PE data with paired ends of varying distance the standard Illumina protocol allows only non-strand-specific paired-end sequencing with a single insert. The insert is normally the stretch of sequence between the paired-end adapters so in your case the insert size would be 250 bp 275 bp reads 100 bp. However the average size of human coding exons is only 160bp.
For instance for a PE150 sequencing run the insert size should be above 300bp. If you have a range of insert sizes you should set it on the mid point of the range. Using the latest Illumina platform the MiSeq paired end reads of 250 and recently even 300 base pairs bp can be obtained.
How to calculate average insert size for paired end reads. It depends on the used Illumina system the used reagents kits and the mode. For Illumina systems DNA insert size range of 200800 bp.
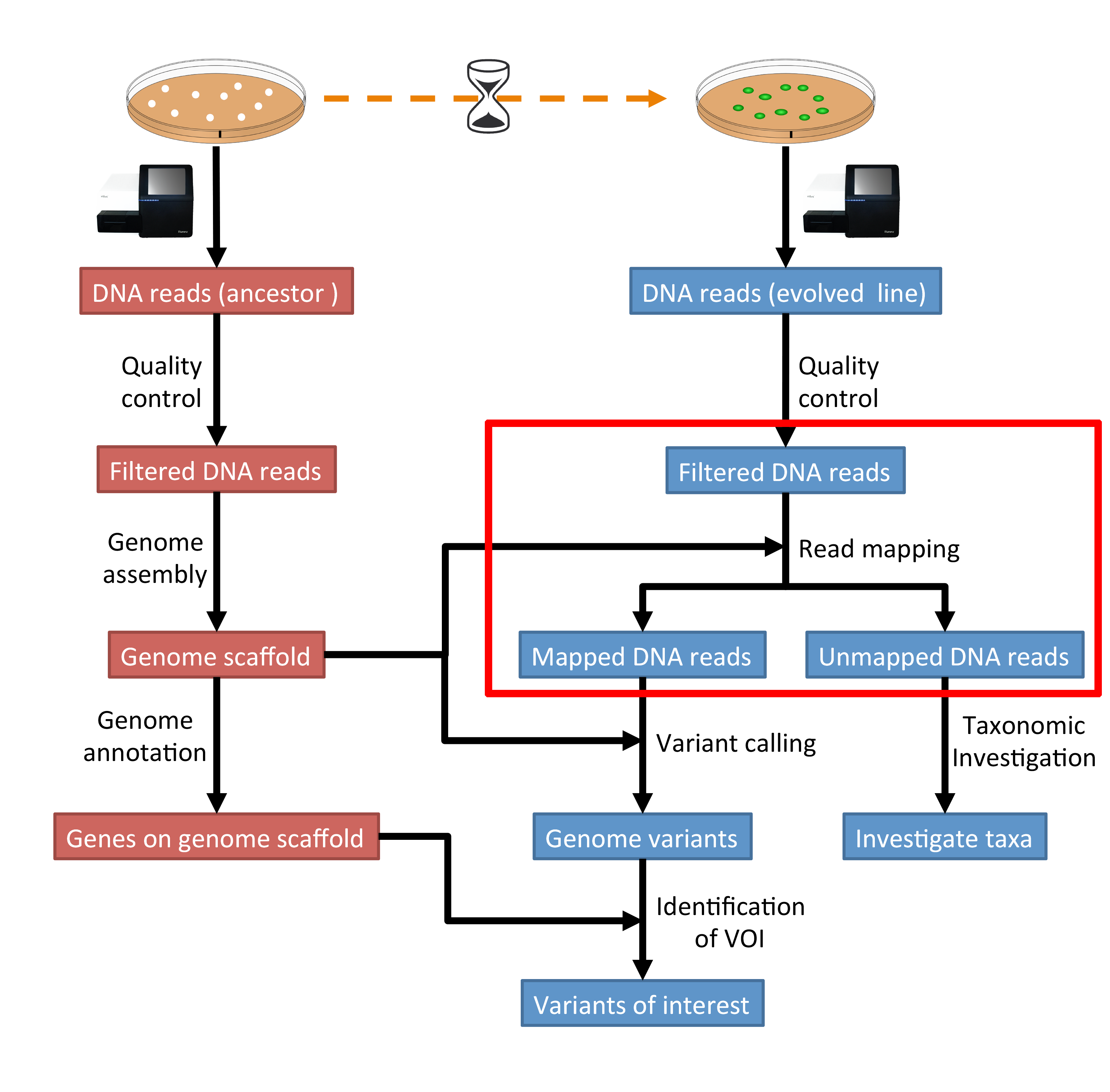
5 Read Mapping Genomics Tutorial 2020 2 0 Documentation
Ppt Sequence Assembly Using Paired End Short Tags Powerpoint Presentation Id 397331
Why Do Illumina Reads All Have The Same Length When Sequencing Differently Sized Fragments
Illumina Short Read Sequencing Lausanne Genomic Technologies Facility
Single End Sequencing Versus Paired End
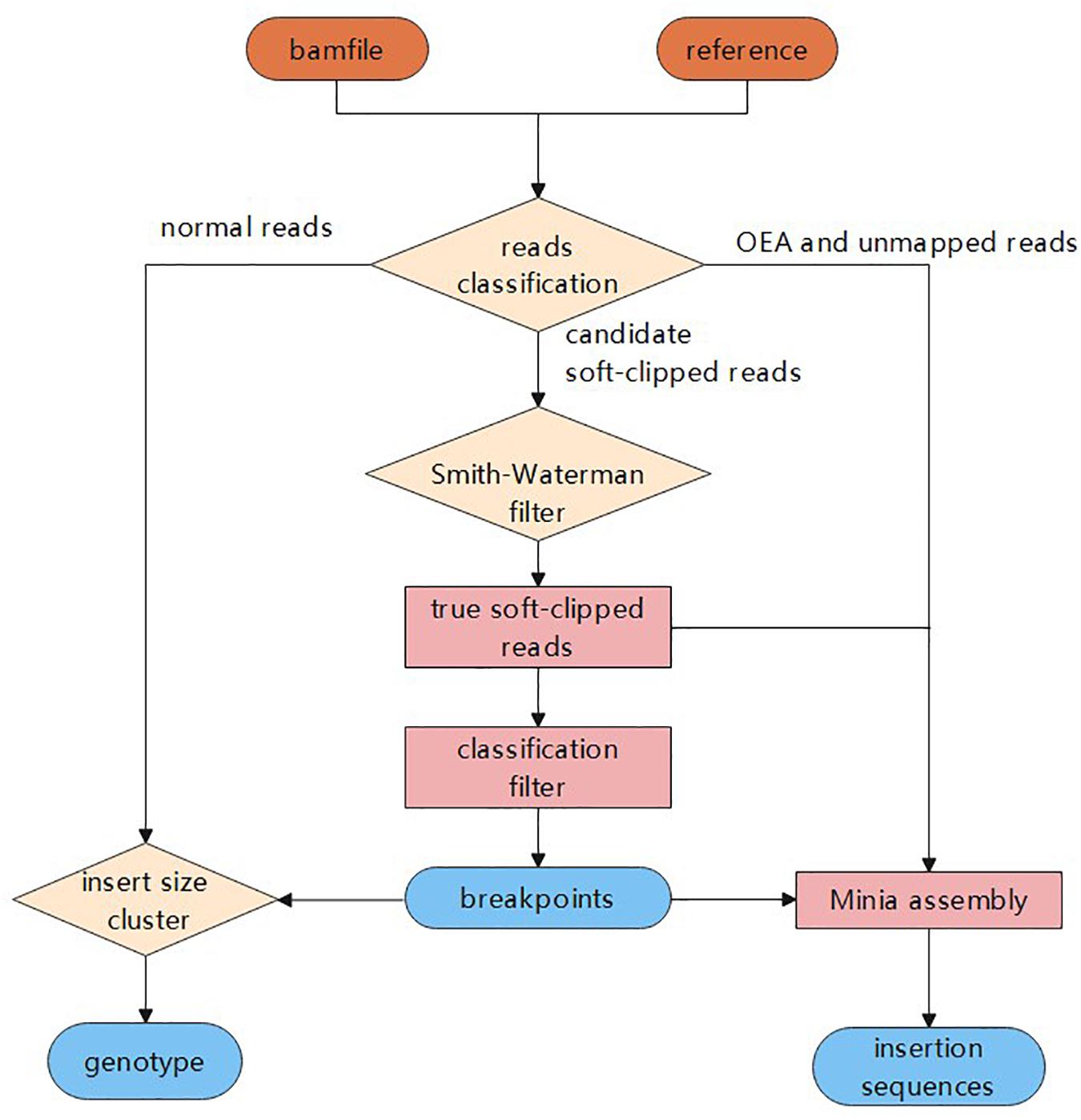
Frontiers Sins A Novel Insertion Detection Approach Based On Soft Clipped Reads
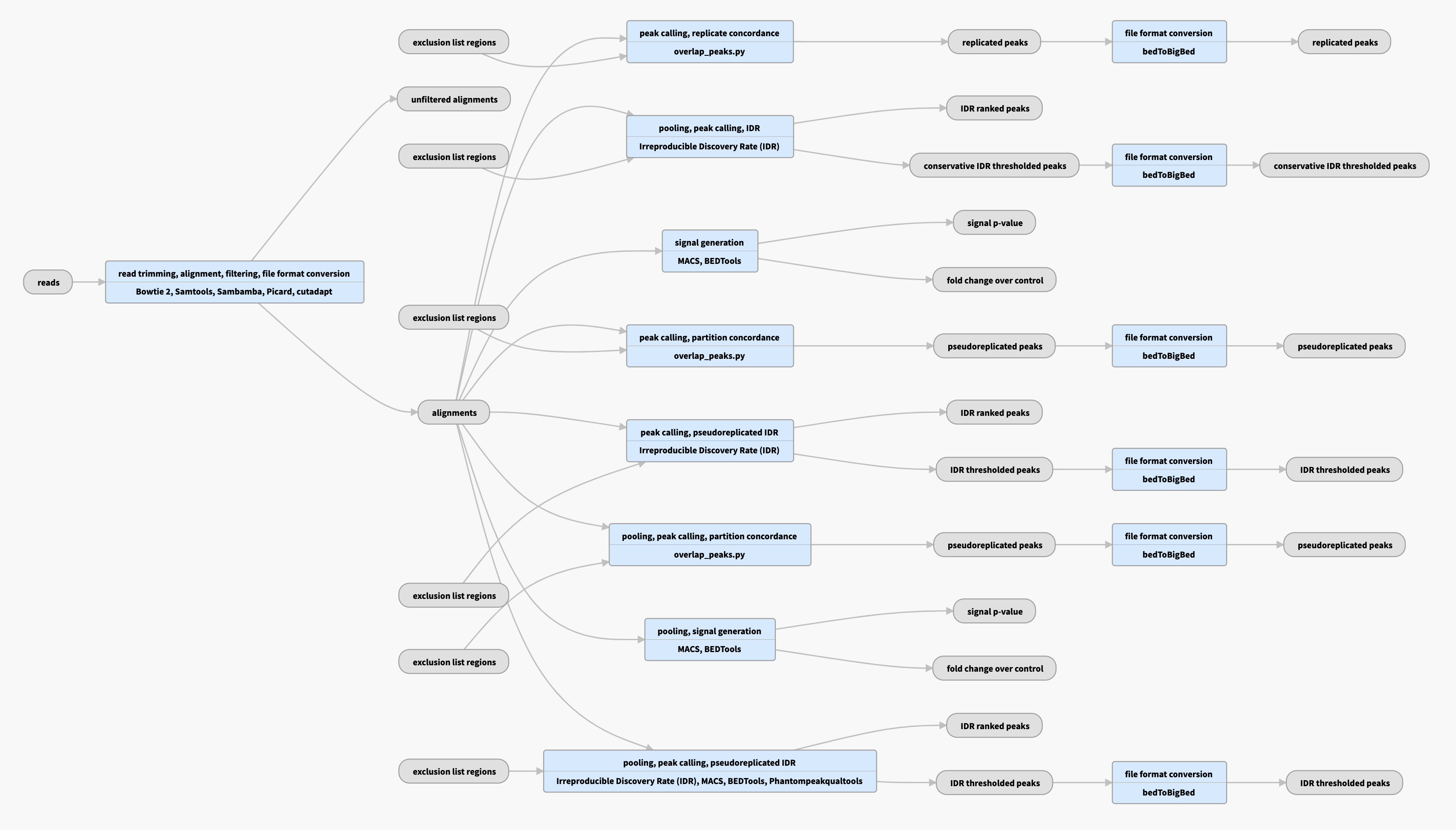
Atac Seq Data Standards And Processing Pipeline Encode

Paired End Pe Sequencing And Insert Size Is Filtering To Increase Download Scientific Diagram

Solved Other Questions 14 If You Have An Insert Size Of Chegg Com
Paired End Sequencing Of Long Range Dna Fragments For De Novo Assembly Of Large Complex Mammalian Genomes By Direct Intra Molecule Ligation Plos One
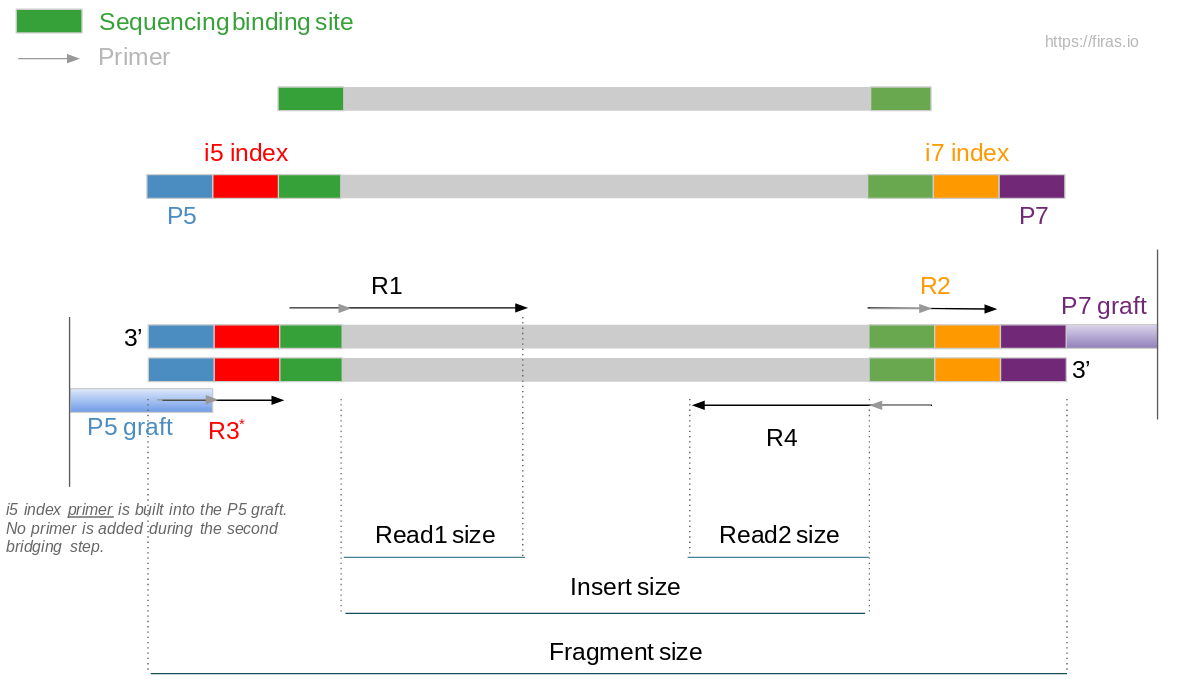
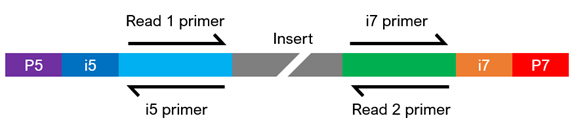
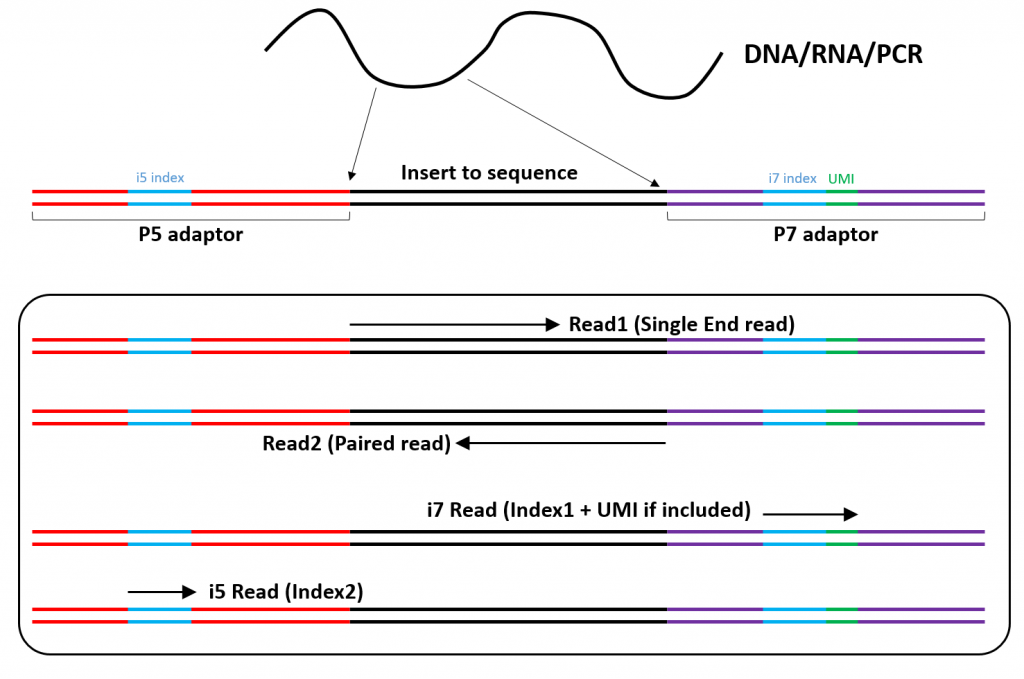
An Overview Of Illumina Multiplexing Firas Sadiyah

Next Generation Sequencing Technology Advances And Applications Sciencedirect
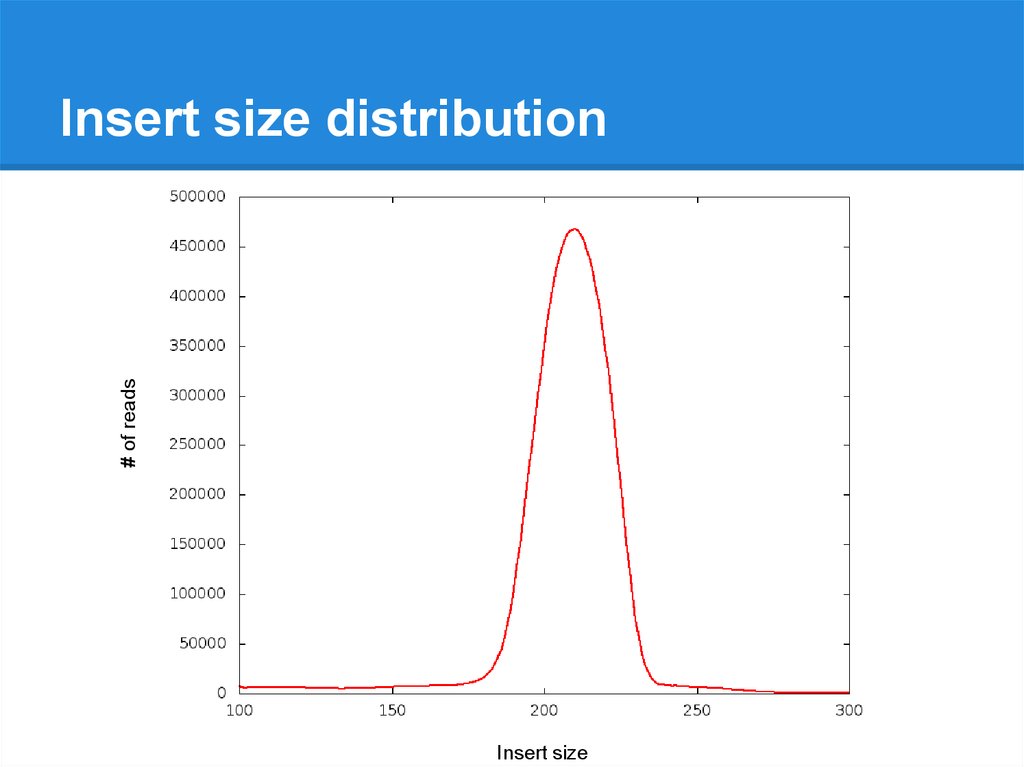
Illumina Data Qc Basic Ngs Tools Prezentaciya Onlajn
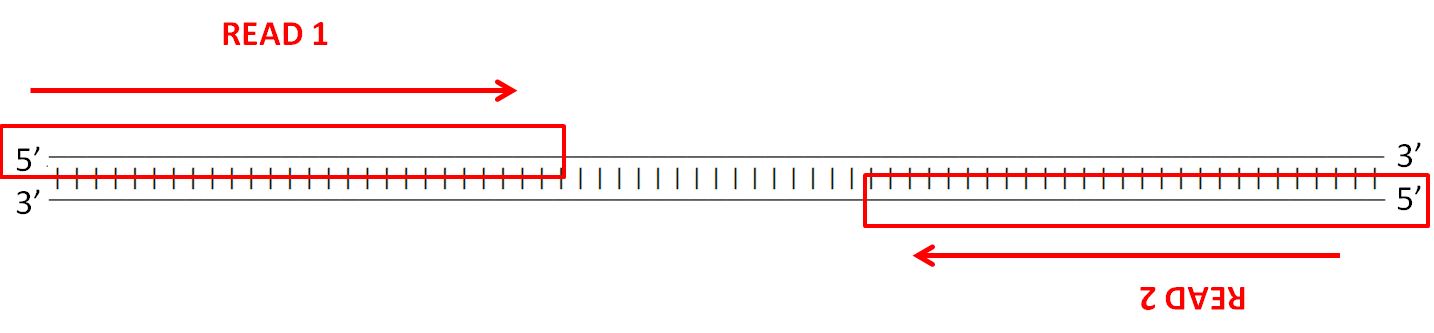
Forward And Reverse Reads In Paired End Sequencing
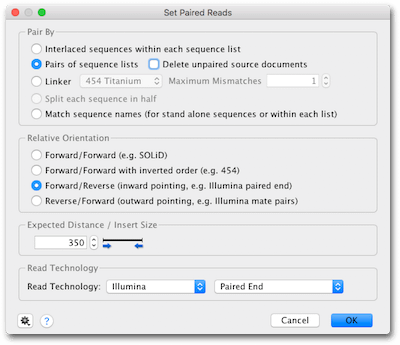
De Novo Assembly Tutorial Geneious Prime
Modification Of The Gs Lt Paired End Library Protocol For Constructing Longer Insert Size Libraries Unt Digital Library
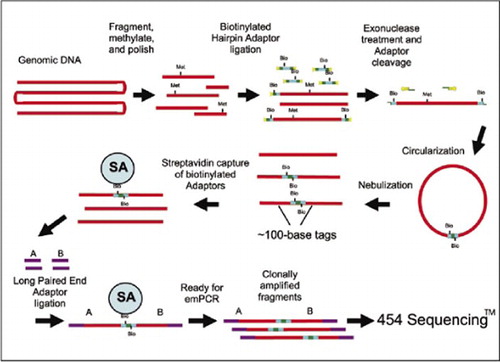
De Novo Assembly And Genomic Structural Variation Analysis With Genome Sequencer Flx 3k Long Tag Paired End Reads Biotechniques